Quick start guide
The easiest way to start importing data into an Ensembl
Dockerised setup
We have incorporated easy-import into to a Dockerised custom Ensembl setup at GenomeHubs.org and this documentation will be fully updated to reflect this approach soon.
Very quick start...
Install docker then:
cd git clone https://github.com/genomehubs/demo demo/demo.sh(best run as UID 1000)
The instructions below will help you get an Ensembl database and website up and running in an afternoon - with four Lepidopteran genomes mirrored from Ensembl Metazoa plus a fresh import of the genome of the winter moth Operophtera brumata direct from publicly hosted .gff and .fasta files.
If you are more interested in importing your own data it's probably still a good idea to check that this example works, then the rest of the documentation will help you get to know the configuration options available and how to customise the .ini files to suit your own data. The sidebar links also provide more detailed descriptions of each of the steps below, and a summary of the relevant configuration options.
If you just want to setup a local Ensembl mirror with existing databases then check out easy mirror, which is included in easy import as a submodule.
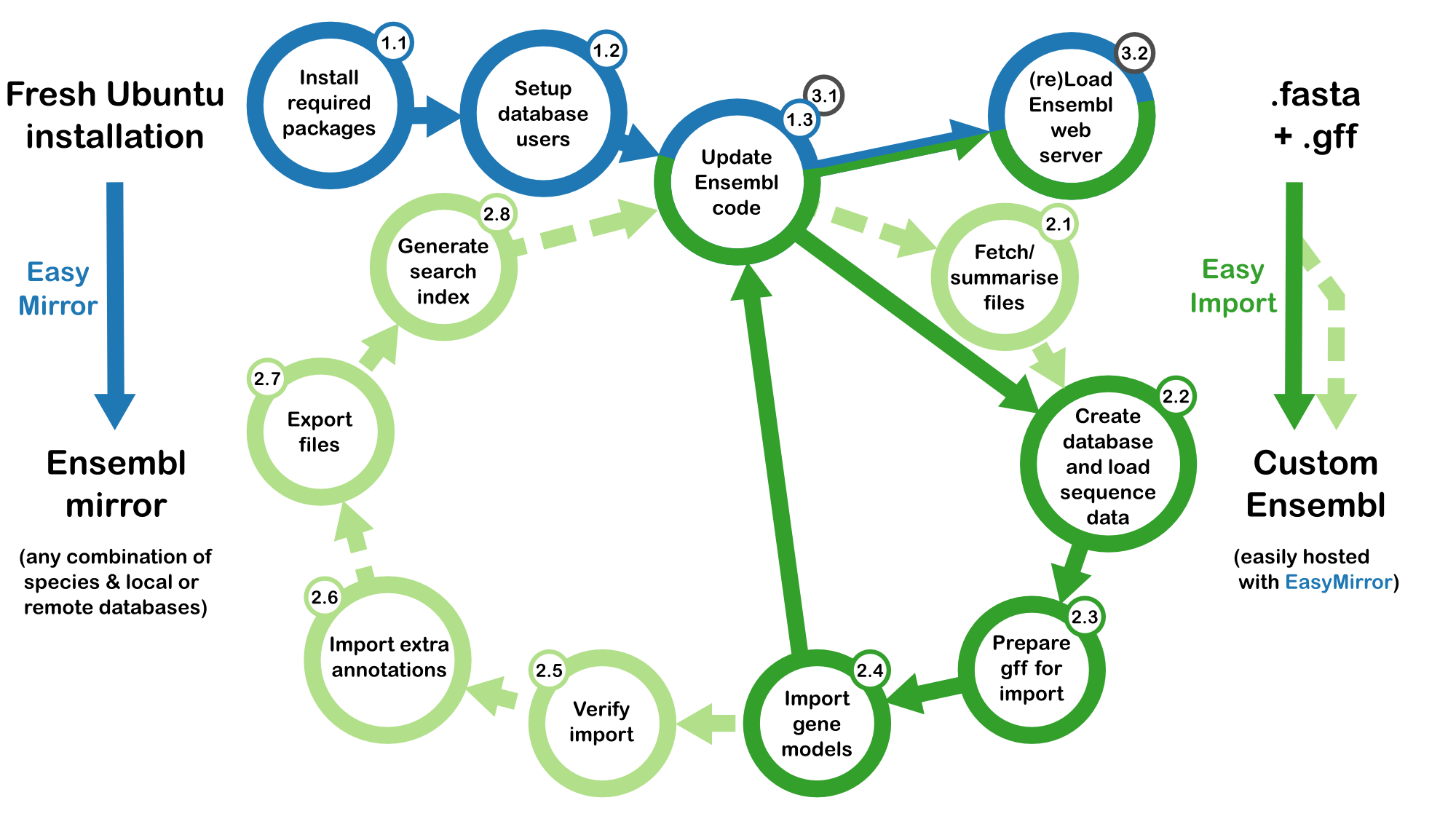
An overview of Ensembl setup with the easy-import pipeline
Stage 1 - Server/database configuration
Using setup.ini/setup-db.ini will setup an Ensembl Metazoa mirror with four Lepidopteran species.
sudo apt-get update
sudo apt-get upgrade
sudo apt-get install git
cd ~
git clone --recursive https://github.com/lepbase/easy-import ei
cd ~/ei/em
sudo ./install-dependencies.sh ../conf/setup.ini
This first step requires root permissions and assumes a fresh install of Ubuntu 14.04 so you might like to read more about what it is doing before running the commands.
cd ~/ei/em
./setup-databases.sh ../conf/setup-db.ini
cd ~/ei/em
./update-ensembl-code.sh ../conf/setup.ini
Stage 2 - Core import
Using core-import.ini will install a new core database for the winter moth Operophtera brumata
mkdir ~/import
cd ~/import
perl ../ei/core/summarise_files.pl ../ei/conf/core-import.ini
cd ~/import
perl ../ei/core/import_sequences.pl ../ei/conf/core-import.ini
perl ../ei/core/import_sequence_synonyms.pl ../ei/conf/core-import.ini
cd ~/import
perl ../ei/core/prepare_gff.pl ../ei/conf/core-import.ini
cd ~/import
perl ../ei/core/import_gene_models.pl ../ei/conf/core-import.ini
cd ~/import
perl ../ei/core/verify_translations.pl ../ei/conf/core-import.ini
cd ~/import
perl ../ei/core/import_blastp.pl ../ei/conf/example.ini ../ei/conf/core-import-extra.ini
perl ../ei/core/import_repeatmasker.pl ../ei/conf/example.ini ../ei/conf/core-import-extra.ini
perl ../ei/core/import_interproscan.pl ../ei/conf/example.ini ../ei/conf/core-import-extra.ini
perl ../ei/core/import_cegma_busco.pl ../ei/conf/example.ini ../ei/conf/core-import-extra.ini
cd ~/import
perl ../ei/core/export_sequences.pl ../ei/conf/core-import.ini
perl ../ei/core/export_json.pl ../ei/conf/core-import.ini
cd ~/import
perl ../ei/core/index_database.pl ../ei/conf/core-import.ini
Stage 3 - Web site configuration
edit setup.ini to add operophtera_brumata_v1_core_31_84_1 to [DATA_SOURCE] SPECIES_DBS
cd ~/ei/em
./update-ensembl-code.sh ../conf/setup.ini
cd ~/ei/em
./reload-ensembl-site.sh ../conf/setup.ini
(optional) Stage 4 - Compara import
Setting up a compara requires the results of a set of analyses in addition to a .ini file similar to compara-import.ini (which contains the configuration used for the lepbase.org compara). Once you have the appropriate files listed in Stage 4 requirements, you will be ready to start importing a a compara.
Updated less than a minute ago
